Protein Production & Characterization
Dr. Anja Schütz
Profil
Our mission is to enable technology development and cutting-edge research by fostering collaboration with other technology platforms and research groups. The PPC Platform offers customized protein production for various downstream applications such as:
- Interaction studies
- High-throughput screening campaigns
- Tool protein production
- Antigen/Antibody production
- Structural biology projects
- Biochemical & biophysical characterization
Next to customized protein production, the PPC Platform is an ideal partner to set up collaborative research projects on topics such as protein engineering and protein characterization. We have a strong interest in studying the structure-function relationship of proteins linked to human diseases. By applying various biochemical and biophysical methods, we delineate the physicochemical properties of the analyzed proteins, a prerequisite to understand their biological function.
The PPC Platform is in close contact with other protein productions sites and partakes in international benchmarking studies initiated through the P4EU network.
Besides being an established partner for educational training of MDC-internal apprentices, the PPC Platform is recruiting bachelor and master students. In the future, we want to promote the success of interdisciplinary PhD projects by offering the possibility of co-supervision of doctoral theses.
Are you interested in a collaboration with us? We are looking forward to your inquiry!
Team
Alumni
We thank our former team members for their active support.
Berger, Ingrid – Technical assistant
Donath, Mandy – Bachelor student
Haake, Fabian – Bachelor student
Lenski, Ulf – Bioinformatician
Kabuß, Loreen-Claudine H. – MDC apprentice
Kuhnke, Alina – MDC apprentice
Kurths, Silke – Technical assistant
Mustroph, Mandy – MDC apprentice
Radusheva, Veselina – Master student
Rossa, Denise – MDC apprentice
Simon, Carolin – Master student
Service & Technology
We have established Standard Operating Procedures (SOPs) and attach particular importance to quality control. Our philosophy is to closely cooperate and actively exchange ideas with our collaborators, since this is essential for successful cooperation.
The following services are offered by the PPC platform:
|
Consulting |
Scientific & technical level |
|---|---|
|
Molecular biology services
|
(1) Construct design (2) Gene cloning (3) Site-directed mutagenesis |
|
Protein production services |
(1) Small-scale expression & solubility screening (2) Scale-up expression (3) Protein purification |
|
Protein characterization services |
(1) Protein stability determination & optimization (2) Protein-ligand interaction studies (ligand = protein, nucleic acid, small molecule) (3) Oligomerization analysis (4) Protein intact mass analysis of peptides and proteins via LC/MS TOF (5) Protein structure determination (including crystallization, X-ray data collection) (6) Folding analysis |
In case you are interested in protein-related services not explicitly listed here, please contact us.
Publications
News
Publications
A: Peer-reviewed publications
E. Specker, S. Matthes, R. Wesolowski, A. Schütz, M. Grohmann, N. Alenina, D. Pleimes, K. Mallow, M. Neuenschwander, A. Gogolin, M. Weise, J. Pfeifer, N. Ziebart, U. Heinemann, J.P. von Kries, M. Nazaré, and M. Bader (2022) Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors. J. Med. Chem., 65, 11126-11149.
Y. Peleg, R. Vincentelli, B.M. Collins, K.-E. Chen, E.K. Livingstone, S.Weeratunga, N. Leneva, Q. Guo, K. Remans, K. Perez, G.E.K. Bjerga, Ø. Larsen, O. Vaněk, O. Skořepa, S. Jacquemin, A. Poterszman, S. Kjær, E. Christodoulou, S. Albeck, O. Dym, E. Ainbinder, T. Unger, A. Schuetz, S. Matthes, M. Bader, A. de Marco, P. Storici, M.S. Semrau, P. Stolt-Bergner, C. Aigner, S. Suppmann, A. Goldenzweig, and S.J.Fleishman (2021) Community-Wide Experimental Evaluation of the PROSS Stability-Design Method. J. Mol. Biol., 433, 166964.
A. Mordhorst, P. Dhandapani, S. Matthes, V. Mosienko, M. Rothe, M. Todiras, J. Self, W.-H. Schunck, A. Schütz, M. Bader, and N. Alenina (2021) Phenylalanine hydroxylase contributes to serotonin synthesis in mice. FASEB J., 35, e21648.
U. Heinemann und A. Schuetz (2019) Structural Features of Tight-Junction Proteins. Int. J. Mol. Sci., 20, 6020.
Q. Ming, Y. Roske, A. Schuetz, K. Walentin, I. Ibraimi, K. Schmidt-Ott, and U. Heinemann (2018) Structural Basis of Gene Regulation by the Grainyhead/CP2 Transcription Factor Family. Nucleic Acids Res., 46, 2082-2095.
S. Quosdorf, A. Schuetz*, and H. Kolodziej* (2017) Different inhibitory potency of oseltamivir carboxylate, zanamivir, and several tannins on bacterial and viral neuraminidases as assessed in a cell-free fluorescence-based enzyme inhibition assay. Molecules, 22, 1989. *shared last authorship
A. Schuetz, V. Radusheva, S.M. Krug, and U. Heinemann (2017) Crystal structure of the tricellulin C-terminal coiled-coil domain reveals a unique mode of dimerization. Ann. N. Y. Acad. Sci., 1405, 147-159.
S. Jennek, S. Mittag, J. Reiche, J.K. Westphal, S. Seelk, M.J. Dörfel, T. Pfirrmann, K. Friedrich, A. Schütz, U. Heinemann, and O. Huber (2017) Tricellulin is a target of the ubiquitin ligase Itch. Ann. N. Y. Acad. Sci., 1397, 157-168.
E. Kowenz-Leutz, A. Schuetz, Q. Liu, M. Knoblich, U. Heinemann, and A. Leutz (2016) Functional interaction of CCAAT/enhancer-binding-protein-α basic region mutants with E2F transcription factors and DNA. Biochim. Biophys. Acta (Gene Regul. Mech.), 1859, 841-847.
Y. Murakawa, M. Hinz, J. Mothes, A. Schuetz, M. Uhl, E. Wyler, T. Yasuda, G. Mastrobuoni, C. C. Friedel, L. Dölken, S. Kempa, M. Schmidt-Supprian, N. Blüthgen, R. Backofen, U. Heinemann, J. Wolf, C. Scheidereit, M. Landthaler (2015) RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-κB pathway. Nat. Commun., 6, 7367.
S. Wagner, A. Schütz, and J. Rademann (2015) Light-switched inhibitors of protein tyrosine phosphatase PTP1B based on phosphonocarbonyl phenylalanine as photoactive phospho-tyrosine mimetic. Bioorg. Med. Chem., 23, 2839-2847.
A. Schuetz, Y. Murakawa, E. Rosenbaum, M. Landthaler, and U. Heinemann (2014) Roquin binding to target mRNAs involves a winged helix-turn-helix motif. Nat. Commun., 5, 5701.
A. Horatscheck, S. Wagner, J. Ortwein, B.G. Kim, M. Lisurek, S. Beligny, A. Schütz, and J. Rademann (2012) Benzoylphosphonate-based photoactive phosphopeptide mimetics for modulation of protein tyrosine phosphatases and highly specific labeling of SH2 domains. Angew. Chem. Int. Ed. Engl., 51, 9441-9447.
F. Kirchner, A. Schuetz, L.-H. Boldt, K. Martens, G. Dittmar, W. Haverkamp, L. Thierfelder, U. Heinemann, and B. Gerull (2012) Molecular insights into arrhythmogenic right ventricular cardiomyopathy caused by plakophilin-2 missense mutations. Circ. Cardiovasc. Genet., 5, 400-411.
F. Mayr, A. Schütz, N. Döge, and U. Heinemann (2012) The Lin28 cold shock domain remodels the terminal loop of pre-let-7 microRNA. Nucleic Acids Res., 40, 7492-7506.
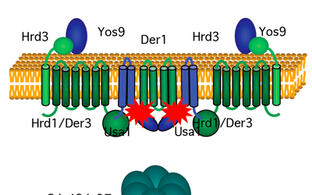
J. Hanna, A. Schütz, F. Zimmermann, J. Behlke, T. Sommer, and U. Heinemann (2012) Structural and biochemical basis of Yos9 protein dimerization and possible contribution to self-association of 3-hydroxy-3-methylglutaryl-coenzyme A reductase degradation ubiquitin-ligase complex. J. Biol. Chem., 287, 8633-8640.
B. von Eyss, J. Maaskola, S. Memczak, K. Möllmann, A. Schuetz, C. Loddenkemper, M.-D. Tanh, A. Otto, K. Muegge, U. Heinemann, N. Rajewsky, and U. Ziebold (2012) The SNF2-like helicase HELLS mediates E2F3-dependent transcription and cellular transformation. EMBO J., 31, 972-985.
D. Busso, Y. Peleg, T. Heidebrecht, C. Romier, Y. Jacobovitch, A. Dantes, L. Salim, E. Troesch, A. Schuetz, U. Heinemann, G.E. Folkers, A. Geerlof, M. Wilmanns, A. Polewacz, C. Quedenau, K. Buessow, R. Adamson, E. Blagova, J. Walton, J.L. Cartwright, L.E. Bird, R.J. Owens, N.S. Berrow, K.S. Wilson, J.L. Sussman, A. Perrakis, and P.H. Celie (2011) Expression of protein complexes using multiple Escherichia coli protein co-expression systems: A benchmarking study. J. Struct. Biol., 175, 159-170.
A. Schuetz, D. Nana, C. Rose, G. Zocher, M. Milanovic, J. Königsmann, R. Blasig, U. Heinemann, and D. Carstanjen (2011) The structure of the Klf4 DNA-binding domain links to self-renewal and macrophage differentiation. Cell. Mol. Life Sci., 68, 3121-3131.
K. Zaragoza, V. Bégay, A. Schuetz, U. Heinemann, and A. Leutz (2010) Repression of transcriptional activity of C/EBPalpha by E2F-dimerization partner complexes. Mol. Cell Biol., 30, 2293-2304.
S.C. Horn, J. Hanna, C., C. Volkwein, A. Schütz, U. Heinemann, T. Sommer, and E. Jarosch (2009) Usa1 functions as a scaffold of the HRD-ubiquitin ligase. Mol. Cell, 36, 782-793.
U.-P. Guenther, L. Handoko, R. Varon, U. Stephani, C.-Y. Tsao, J.R. Mendell, S. Lützkendorf, C. Hübner, K. von Au, S. Jablonka, G. Dittmar, U. Heinemann, A. Schuetz*, and M. Schuelke* (2009) Clinical variability in distal spinal muscular atrophy type 1 (DSMA1): determination of steady-state IGHMBP2 protein levels in five patients with infantile and juvenile disease. J. Mol. Med., 87, 31-41. *shared corresponding authorship
S. Gräslund et al. (2008) Protein production and purification. Nat. Methods, 5, 135-146.
B: Acknowledgements
J. Kemna, E. Gout, L. Daniau, J. Lao, K. Weißert, S. Ammann, R. Kühn, M. Richter, C. Molenda, A. Sporbert, D. Zocholl, R. Klopfleisch, H. Lortat-Jacob, P. Aichele, T. Kammertoens, and Blankenstein (2023) IFNγ binding to extracellular matrix prevents fatal systemic toxicity. Nature Immunology, 24, 414-422.
M. Broto, M.M. Kaminski, C. Adrianus, N. Kim, R. Greensmith, S. Dissanayake-Perera, A.J. Schubert, X. Tan, H. Kim, A.S. Dighe, J.J. Collins, and M.M. Stevens (2022) Nanozyme-catalysed CRISPR assay for preamplification-free detection of non-coding RNAs. Nature Nanotechnology, 17, 1120-1126.
A.C. McGarvey, W. Kopp, D. Vucicevic, K. Mattonet, R. Kempfer, A. Hirsekorn, I. Bilic, M. Gil, A. Trinks, A.M. Merks, D. Panáková, A. Pombo, A. Akalin, J.P. Junker, D.Y.R. Stainier, D. Garfield, U. Ohler, and S.A. Lacadie (2022) Single-cell-resolved dynamics of chromatin architecture delineate cell and regulatory states in zebrafish embryos. Cell Genomics, 2, 100083.
U. Zinnall, M. Milek, I. Minia, C.H. Vieira-Vieira, S. Müller, G. Mastrobuoni, O.G. Hazapis, S. Del Giudice, D. Schwefel, N. Bley, F. Voigt, J.A. Chao, S. Kempa, S. Hüttelmaier, M. Selbach, and M. Landthaler (2022) HDLBP binds ER-targeted mRNAs by multivalent interactions to promote protein synthesis of transmembrane and secreted proteins. Nature Communications, 13, 2727.
W. Winick-Ng, A. Kukalev, I. Harabula, L. Zea-Redondo, D. Szabó, M. Meijer, L. Serebreni, Y. Zhang, S. Bianco, A.M. Chiariello, I. Irastorza-Azcarate, C.J. Thieme, T.M. Sparks, S. Carvalho, L. Fiorillo, F. Musella, E. Irani, E.T. Triglia, A.A. Kolodziejczyk, A. Abentung, G. Apostolova, E.J. Paul, V. Franke, R. Kempfer, A. Akalin, S.A. Teichmann, G. Dechant, M.A. Ungless, M. Nicodemi, L. Welch, G. Castelo-Branco, and A. Pombo (2021) Cell-type specialization is encoded by specific chromatin topologies. Nature, 599, 684-691.
E. Dias D'Andréa, Y. Roske, G.A.P. de Oliveira, N. Cremer, A. Diehl, P. Schmieder, U. Heinemann, H. Oschkinat, and J. Ricardo Pires (2020) Crystal structure of Q4D6Q6, a conserved kinetoplastid-specific protein from Trypanosoma cruzi. Journal of Structural Biology, 211, 107536.
M. Zech, R. Jech, S. Boesch, M. Škorvánek, S. Weber, M. Wagner, C. Zhao, A. Jochim, J. Necpál, Y. Dincer, K. Vill, F. Distelmaier, M. Stoklosa, M. Krenn, S. Grunwald, T. Bock-Bierbaum, A. Fecíková, P. Havránková, J. Roth, I. Príhodová, M. Adamovicová, O. Ulmanová, K. Bechyne, P. Danhofer, B. Veselý, V. Han, P. Pavelekova, Z. Gdovinová, T. Mantel, T. Meindl, A. Sitzberger, S. Schröder, A. Blaschek, T. Roser, M.V. Bonfert, E. Haberlandt, B. Plecko, B. Leineweber, S. Berweck, T. Herberhold, B. Langguth, J. Švantnerová, M. Minár, G.A. Ramos-Rivera, M.H. Wojcik, S. Pajusalu, K. Õunap, U.A. Schatz, L. Pölsler, I. Milenkovic, F. Laccone, V. Pilshofer, R. Colombo, S. Patzer, A. Iuso, J. Vera, M. Troncoso, F. Fang, H. Prokisch, F. Wilbert, M. Eckenweiler, E. Graf, D.S. Westphal, K.M. Riedhammer, T. Brunet, B. Alhaddad, R. Berutti, T.M. Strom, M. Hecht, M. Baumann, M. Wolf, A. Telegrafi, R.E. Person, F.M. Zamora, L.B. Henderson, D. Weise, T. Musacchio, J. Volkmann, A. Szuto, J. Becker, K. Cremer, T. Sycha, F. Zimprich, V. Kraus, C. Makowski, P. Gonzalez-Alegre, T.M. Bardakjian, L.J. Ozelius, A. Vetro, R. Guerrini, E. Maier, I. Borggraefe, A. Kuster, S.B. Wortmann, A. Hackenberg, R. Steinfeld, B. Assmann, C. Staufner, T. Opladen, E. Ružicka, R.D. Cohn, D. Dyment, W.K. Chung, H. Engels, A. Ceballos-Baumann, R. Ploski, O. Daumke, B. Haslinger, V. Mall, K. Oexle, and J. Winkelmann (2020) Monogenic variants in dystonia: an exome-wide sequencing study. Lancet Neurology, 19, 908-918.
D. Sundaravinayagam, A. Rahjouei, M. Andreani, D. Tupina, S. Balasubramanian, T. Saha, V. Delgado-Benito, V. Coralluzzo, O. Daumke, and M. Di Virgilio (2019) 53BP1 supports immunoglobulin class switch recombination independently of its DNA double-strand break end protection function. Cell Reports, 28, 1389-1399.
H. Behrmann, A. Lürick, A. Kuhlee, H. Kleine Balderhaar, C. Bröcker, D. Kümmel, S. Engelbrecht-Vandré, U. Gohlke, S. Raunser, U. Heinemann, and C. Ungermann (2014) Structural identification of the VPS18 β-propeller reveals a critical role in the HOPS complex stability and function. Journal of Biological Chemistry, 289, 33503-33512.
F. Mayr and U. Heinemann (2013) Mechanisms of Lin28-mediated miRNA and mRNA regulation: a structural and functional perspective. International Journal of Molecular Sciences, 14, 16532-16553.
S. Boivin and D.J. Hart (2011) Interaction of the influenza A virus polymerase PB2 C-terminal region with importin a isoforms provides insights into host adaptation and polymerase assembly. Journal of Biological Chemistry, 286, 10439-10448.
H. Striegl, M.A. Andrade-Navarro, and U. Heinemann (2010) Armadillo motifs involved in vesicular transport. PLoS ONE, 5, e8991.
C: Structure Gallery
Below, you find our crystal structure depositions into the Protein Data Bank.
7ZIF, 7ZIG, 7ZIH, 7ZII, 7ZIJ, 7ZIK, 6EKU, 6EKS, 5N7H, 5N7I, 5N7K, 5MR7, 5MPH, 5MPI, 5MPF, 4ULW, 3ZM0, 3ZM1, 3ZM2, 3ZM3, 4A75, 4A76, 4ALP, 3ULJ, 4A4I, 3TT9, 2YMA, 2WBS, 2WBU
D: Patents
M. Bader, E. Specker, S. Matthes, A. Schütz, K. Mallow, M. Grohmann, M. Nazaré. Xanthine derivatives, their use as a medicament, and pharmaceutical preparations comprising the same. Europe Patent WO/2018/019917 filed 28 Jul 2016, and issued 1 Feb 2018.
M. Bader, E. Specker, S. Matthes, A. Schütz, K. Mallow, M. Grohmann, M. Nazaré. Xanthine derivatives, their use as a medicament, and pharmaceutical preparations comprising the same. Europe Patent WO/2016/135199 filed 24 Feb. 2015, and issued 1 Sept. 2016.