Izsvák Lab
Mobile DNA
Profile
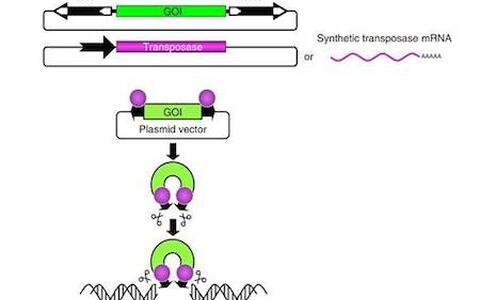
The two-component Sleeping Beauty (SB100X)-based non-viral vector system
Transposable elements (TEs) are discrete segments of DNA that have the distinctive ability to move from one genetic location to another in a genome. Although, large fractions of genomes can be composed of TE-derived sequences (~55% of the human genome), the vast majority of these elements are not essential to the host cell, and often referred as ’junk DNA’ As an attempt to turn “junk” into treasure, the molecular reconstruction of ancient transposable elements, including Sleeping Beauty represents a milestone in applying transposition-mediated gene delivery in vertebrate species. My focus is to understand the basic mechanism of transposition and the interplay between TEs and host cells in mammals. My team integrates basic knowledge with technologies that are revolutionizing genomic manipulations in vertebrate species, including gene therapy, transgenesis, cancer research and functional genomics. My studies also focuses on evolutionary processes that seem to “recycle” inactive TEs and co-opt them for novel cellular function, and argue against the dogma that views them exclusively as parasites or “junk” DNA.
Team
Research
Aims and plan for the next 5-year term
The evolutionary success of transposable elements (TEs) is powerfully underscored by the finding that about 60% of the human genome is TE-derived. TE-associated activities are elevated following large environmental changes, and could be beneficial for a population. Nevertheless, they might be deleterious for an individual by promoting disease. The best-characterized scenarios are directly associated with de novo transposition or recombination events provoked by active elements (e.g. LINE-1). Although, a very small proportion of TEs can potentially encode protein(s), a plethora of data indicate that even transpositionally defective copies can be actively transcribed, pointing to a retained transcriptional regulatory function for thousands of TEs. In fact, there are at least four reasons to believe that the impact of TEs on human physiology is deeply underestimated: (i) the abundance of TEs in the human genome (ii) the ability of TE sequences to create novel transcripts, (iii) the relationship between TEs and RNA-based gene regulation, and (iv) their inducibility by stress. Therefore, significant efforts required focusing to decipher this so-far neglected area of medical research. Thus, there is an urgent need to revisit and understand the contribution of TEs to genomic instability and human disease. A better understanding of the association between stress and TE-induced molecular evolution would allow us to work out strategies to cure a number of diseases. We recently set out to investigate the impact of TE-derived activities on the human genome in general and on disease mechanisms in particular, based on the central premise that some of these activities are stress-induced. In parallel, we aim at translating experience accumulated in TE research to cutting-edge technologies. Our goal is to integrate basic knowledge with technologies that are revolutionizing genomic manipulations in vertebrate species, including gene therapy, transgenesis, cancer research and functional genomics. The anticipated output of our research is a refined understanding of the consequences of environmental stress on our genome mediated by TE-derived sequences.