Mittnenzweig Lab
Computational and developmental biology
Profil
Our broad goal is to develop quantitative models of dynamical biological processes in development and disease integrating data from multiomic and imaging technologies. In collaboration with experimental groups, we study how cells regulate the expression of their genes, how they communicate with their neighbors and how structure emerges at the tissue level from the interaction of many cells. A focus of the lab is the fascinating process of embryonic development, during which an entire animal forms from a single egg cell. Using computational tools, we reconstruct the trajectories in space and time that cells take during their journey, with the ultimate goal of understanding how, where and when cell fate decisions are made. Through in vitro models such as gastruloids or embryoid bodies we can reconstitute in vivo development and test and dissect the effect of specific factors on cellular and tissue-wide differentiation. Along the way, we are developing computational methods and models for new genomic data modalities and integrate the large amount of available single-cell and spatial genomic data.
Team
Research
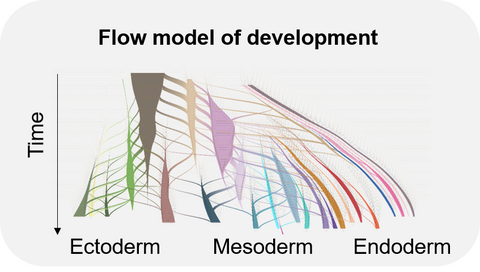
1) Reconstructing cellular trajectories during development
Throughout development, cells continuously divide and differentiate leading to more and more specialized cell types. Understanding the mechanisms underlying these cell fate decisions is challenging because of the interplay of various intrinsic (transcription factors, epigenetic changes) and extrinsic (signaling molecules) factors. In the context of gastrulation, we recently inferred precise differentiation flows and lineage specification dynamics for every final transcriptional state using a network flow model approach. With the increasing availability of large-scale single-data covering also later developmental stages, we are working towards building spatiotemporal flow models of later development and extending this approach to other data modalities (e.g. lineage, spatial, epigenomic information).
2) From cells to tissues
Cell-cell communication is essential for cellular specification and tissue patterning throughout development and in adult life. In particular, cells and tissues reuse a small number of signaling pathways such as Wnt, Bmp, Nodal, Notch and Shh in diverse developmental contexts. While the components of each pathway are well-known, a mechanistic understanding of how these pathways are employed during development to ensure robust cellular differentiation and pattern formation is currently missing. In the coming years, we want to develop quantitative models that will explain how these pathways work together to drive cellular differentiation and spatial structure. The starting point is a quantitative description of the phenomenology of cellular differentiation that we have developed in the last years. Developmental signaling pathways are also involved in many diseases and a better understanding of their functionality during normal development might also present new opportunities for understanding their dynamics in disease processes.
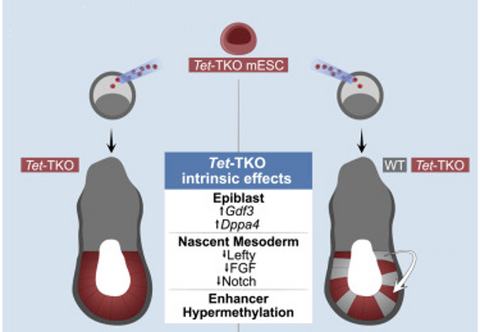
3) Understanding the effect of epigenetic changes during development
Early mammalian embryonic development involves dramatic changes in the epigenetic state of its cells. In particular, DNA methylation rises from almost no methylation in inner cell mass cells to very high methylation levels in epiblast cells at the onset of gastrulation. The three TET proteins can actively demethylate DNA and mouse embryos deficient of TET die shortly after gastrulation. Revisiting this mutant phenotype using temporal single-cell atlases from embryos with partial or complete mutant contributions allowed to quantify and dissect early intracellular effects of the TET KO from secondary tissue-wide repercussions. Strikingly, when developing within a wild-type embryo, Tet-mutant cells retain near-complete differentiation potential, whereas full-embryo mutants fail to generate ectoderm and most embryonic mesoderm lineages. Moving forward, we want to understand better the contribution of intra- and intercellular changes on cell fate decisions and develop temporal models of the single-cell epigenome during development.