Piazza Lab
Allosteric Proteomics
Profile
Binding of small molecules to proteins can modulate the metabolic status of the cells and their gene expression dynamics. All these interaction events between protein and metabolites produce allosteric effects, which means that they regulate protein activity as a consequence of a protein structural change.
We study how cells adapt to different metabolite compounds at the molecular level, focusing on human diseases where the balance between cellular proliferation and differentiation is impaired, due to a loss of regulation of the cellular chemical reactions. Our research approach integrates multiple system biology approaches with a particular focus on novel proteomics methods and genomics.
For more information about our research vision and technological interests visit the laboratory website linked in the social media channels Twitter and LinkedIN, as it is more up to date and informal than this institutional webpage.
Team
Group Leader
Senior Scientists
Dr. Claudio Bassot (Senior Bioinformatician)
Dr. Tomas Koudelka (Laboratory Manager and mass spectrometry scientist)
Postdocs
PhD students
Secretary
For more information about our team interests visit the laboratory website linked in the social media channels Twitter and LinkedIN, as it is more up to date and informal than this institutional webpage.
Research
OUR VISION
From a mechanistic point of view cells can be seen as integrated devices composed of a network of interactions among different macromolecules and small molecules (proteins, nucleic acids and metabolites). Most of them are protein molecular machines that carry out specific tasks, such as building structures and catalyse reactions. In our team we try to understand these complex networks and how proteins are activated. This is also important to design strategies to correct these processes when they get out of control in diseases.
During the last decade a new discipline of biology called proteomics has changed the paradigm of the methods used to study biological mechanisms. Rather than recapitulating the (bio)chemistry of single reactions reconstructed in a test tube, proteomics aims to capture a snapshot of all biological processes and molecular event occurring in vivo by profiling the levels of proteins present in the cell. Unfortunately, knowing protein abundances is not often sufficient to explain phenotypes. This is because the activity of all proteins depends not only on their abundance but also on their 3D structure. Ultimately, all factors that can modify protein’s shapes, most relevantly molecular interactions with metabolites, drugs. nucleic acids and other proteins are important for their function. Thus, a systematic and comprehensive understanding of biological processes has to take into account all these heterogenous interactions.
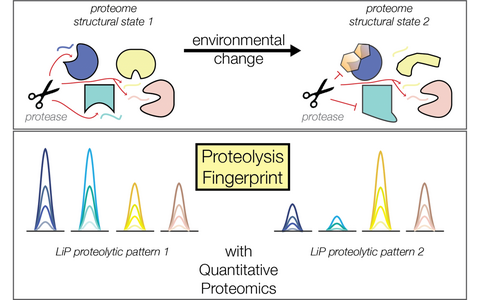
Probing structural states of the proteome combing limited proteolysis and mass spectrometry (LiP-MS concept).
Guided by the paradigm that a change in activity generally correspond to a change in structure, we use proteomics to study protein structural changes associated to many different types of molecular cues. For instance:
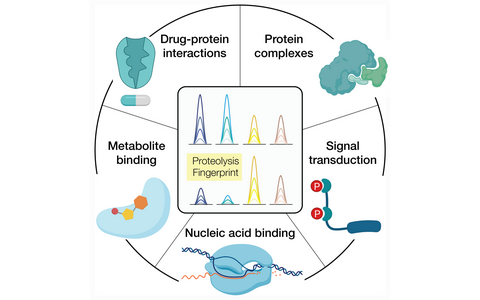
- Assembly-disassembly of protein complexes
- The effects of phosphorylation on protein function
- Interactions of protein to metabolites
- Identification of protein drug targets in vitro and in vivo
- Dynamic interactions of proteins with nucleic acids
- Protein stability changes due to mutations or aggregation
Different kinds of cellular social interactions that can be investigated with the LiP-MS technology.
We are also very interested in advancing further the field of structural systems biology by improving new workflows. With the multidisciplinary research and collaborative atmosphere of the MDC, we aim to combine these state-of-the-art proteomics methods with genomics, metabolomics, stem cell biology and computational biology. Our general goal Is to advance the general knowledge of basic research and make a direct impact on topics related to human health. We are in the right environment to do so.
OUR GOALS
Different environmental cues such as stress, nutrients or drugs, trigger rapid adaptive responses that allow to maintain cellular homeostasis. One of the fastest cellular responses to the environment is the binding of small molecules to proteins. These molecular interactions produce allosteric effects, which means that they trigger a variation of protein activity as a consequence of a structural conformational changes that occurs instantly. Allosteric interactions are thus essential for life and can modulate both the metabolic status of the cells and gene expression.
We study how cells adapt to different metabolite compounds at the molecular level, focusing on the regulation of the balance between cellular proliferation and terminal differentiation in stem cells. Metabolite levels can control both the inherent proliferative properties of stem cells in normal conditions but also their uncontrolled proliferation, which may lead to diseases like cancer.
In cancer diseases leukaemia the balance between cellular proliferation and differentiation is impaired. Our team is interested in understanding how this happens at the molecular level. We will use multiple system biology approaches with a particular focus on novel proteomics methods, metabolomics and genomics to study:
- The principles and pervasiveness of metabolite regulation of chromatin.
- How metabolites control stem cells fate in leukaemia.
- How to efficiently modulate the chromatin-metabolite network with drugs.
To tackle these questions we need advanced bioinformatics and a structural-system biology prospective. We will integrate these different levels of gene regulation into predictive models that elucidate their function in collaboration with other research groups at the MDC and with the Berlin institute of medical system biology.