Robson Lab
Chromatin (dys)function in disease
Profile
The identity and function of every cell in the body is defined by the genes it activates. Cells achieve such control by packaging their genome into chromatin, a sophisticated 3D structure that encodes normal gene activities at multiple layers (Robson et al, 2019; Ringel et al, 2022). Disruptions to chromatin are therefore a major driver of disease.
Understanding how chromatin’s regulatory layers collectively define gene activity or drive human pathology are thus central questions in the Robson lab. To pursue this, we implement and develop single cell methods to explore how chromatin features form, control gene activity, and mechanistically cause disease. Moreover, we study these normal and pathogenic processes in the actual tissue environments where gene regulation actually operates.
Our experimental and computational approaches are interdisciplinary at all scales, combining in vivo and organoid models with genetics, single cell genomics, and imaging. We seek for the lab to match this by creating an inclusive environment that supports diverse backgrounds, temperaments, goals, and opinions.
Team
Research
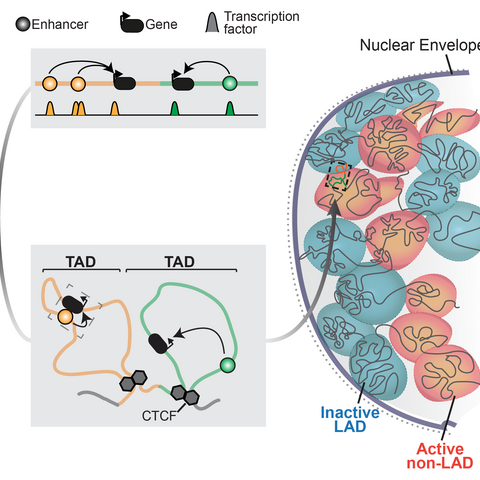
Summary of chromatin regulation of genes (adapted from Robson et al, 2019)
Gene regulation is essential for life and is controlled at multiple layers. Instructions activate genes in specific tissues are scattered throughout surrounding chromatin in enhancer sequences that recruit epigenetic regulators (Long et al, 2016). 3D chromatin structure then conveys these regulatory activities by physically connecting enhancers with their correct target genes (Andrey et al, 2017). Finally, the genome’s interactions with the nuclear envelope further structures chromatin to isolate and repress inactive genes (Robson et al, 2019). Together, these chromatin features define a gene’s regulatory landscape and, as we recently showed, can be assembled in many combinations to uniquely control any locus (Ringel et al, 2022).
Our lab now aims to mechanistically understand how these regulatory landscape components interact to collectively control transcription and how disrupting this drives disease. We do this using a truly interdisciplinary approach, combining wet lab and computational approaches with mice, organoids, single cell multi-omics and microscopy.
In particular, we are fascinated by how examining unique biological phenomena with new technologies and perspectives can reveal entirely new regulatory principles. We currently tackle three main questions;
1) How are regulatory landscapes turned on and off?
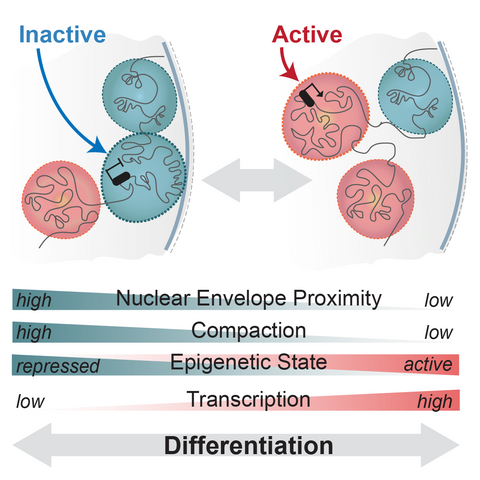
Schematic of regulatory landscapes reconfiguration in development (adapted from Robson et al, 2019)
Regulatory landscapes transition between active and inactive states as cells differentiate or respond to cues. However, the step-wise series of events that enable this remain unknown. Do landscapes decompact and release from the nuclear envelope before their genes can be activated? How and when does the binding of transcription factors enable this? What happens if these transitions are synthetically disrupted? We aim to uncover these principles in vivo using CRISPR genome engineering and single cell multi-omics.
2) How are landscapes disrupted in disease?
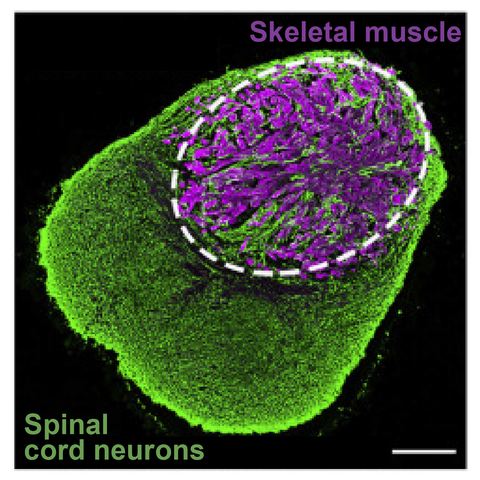
Example human neuromuscular organoids that model neurons and innervated myofibers (adapted from Martins et al, 2020)
The functions of many components of regulatory landscapes are poorly understood and so it is unclear how their disruption drives disease. For example, mutations in the nuclear envelope proteins that attach chromatin cause “laminopathies” ranging from premature aging to muscular dystrophy. How does this disrupt regulatory landscapes and their components to promote disease? Can this be reversed? We will address this by combining organoid and mouse models of laminopathies with novel single cell genomics and imaging.
3) How are chromatin & landscape components repurposed?
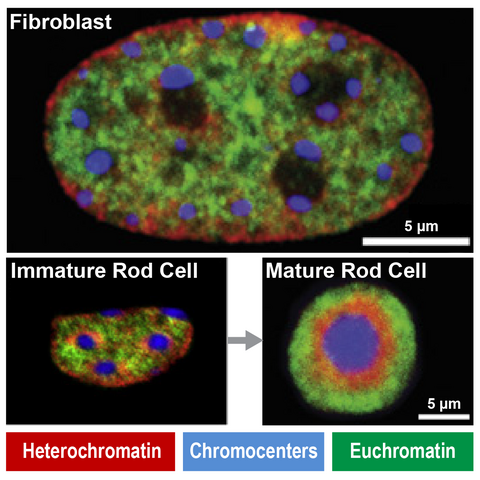
Immunofluorescence show unique heterochromatin in mouse rod photoreceptors that are adapted for nocturnal vision (adapted from Solovei et al, 2007)
Fundamental components of chromatin can be modified to serve new functions in evolution. For example, in nocturnal mammals like mice rod cell photoreceptors severe all nuclear envelope-heterochromatin interactions. Accordingly, inactive regulatory landscapes become uniquely released into the nuclear interior to improve the retina’s light sensitivity. How does heterochromatin release occur in rods over time? What consequences does “inversion” or its ectopic reversal have for regulatory landscape function? Does inappropriate inversion cause human disease? We address this by combining mouse models and retinal organoids with single cell genomics, imaging and proteomics.
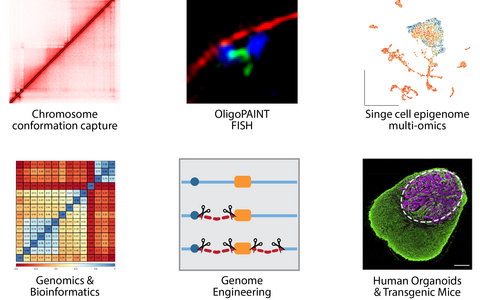
“Our toolkit”
Publications
News
Jobs
Join us! We are always looking for passionate new lab members (Postdocs, PhD and Master’s students). Please drop Mike an email to learn more.